Leveraging pleiotropy identifies a novel sleep apnea locus
27 Dec 2022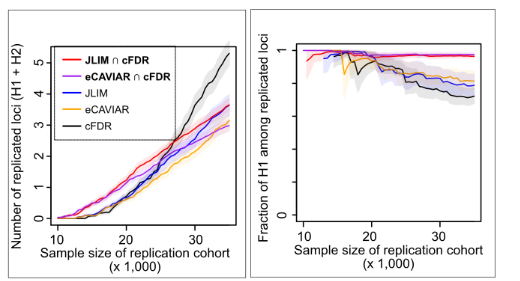 Small-scale studies often have inadequate power to discover new genetic associations. Traits that are burdensome or expensive to phenotype, rare diseases that are hard to sample and diseases that affect under-represented populations could all lead to such underpowered genetic studies. Here, we present a strategy to leverage pleiotropy between traits to discover new loci in underpowered traits. Specifically, we employ an approach that we call “consensus pleiotropy,” which combines a colocalization test with a locus-level test of pleiotropy. In simulations, we show that this approach is highly selective for identifying true pleiotropy driven by the same causative variant, thereby improves the chance to replicate the associations in underpowered validation cohorts. Using Obstructive Sleep Apnea (OSA) as an examplar, we show that we can identify and replicate a novel OSA-association in PRIM1 (DNA primase subunit 1) locus in small genetic cohorts. See Chun and Akle et al. PLOS Genet 2022 for more details of our work.
Small-scale studies often have inadequate power to discover new genetic associations. Traits that are burdensome or expensive to phenotype, rare diseases that are hard to sample and diseases that affect under-represented populations could all lead to such underpowered genetic studies. Here, we present a strategy to leverage pleiotropy between traits to discover new loci in underpowered traits. Specifically, we employ an approach that we call “consensus pleiotropy,” which combines a colocalization test with a locus-level test of pleiotropy. In simulations, we show that this approach is highly selective for identifying true pleiotropy driven by the same causative variant, thereby improves the chance to replicate the associations in underpowered validation cohorts. Using Obstructive Sleep Apnea (OSA) as an examplar, we show that we can identify and replicate a novel OSA-association in PRIM1 (DNA primase subunit 1) locus in small genetic cohorts. See Chun and Akle et al. PLOS Genet 2022 for more details of our work.