. We are interested in the development of novel computational methods for integrative genetic analysis and their application to rare and common pulmonary genetic disorders. Chun Lab is supported by NHLBI (1R56HL171620).
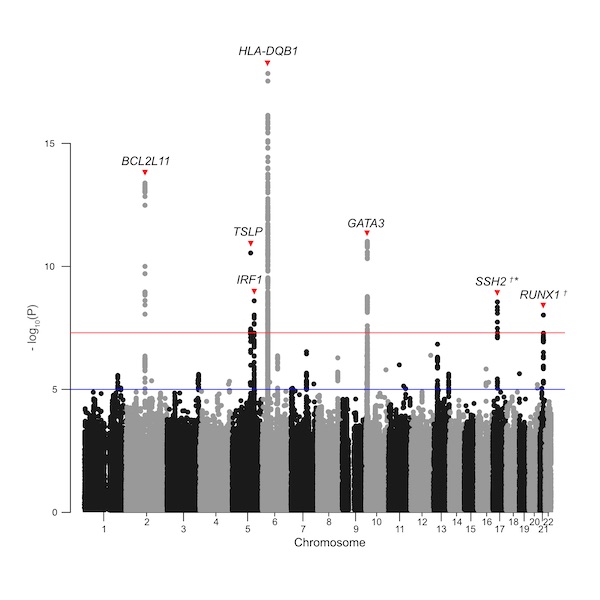 Eosinophilic granulomatosis with polyangiitis (EGPA) is a rare anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis characterized by eosinophilia and severe asthma. We conducted a Genome-wide Association Study (GWAS) of EGPA in a total of 928 cases and 11,266 controls of European ancestry. We present our findings at the annual meeting of American Thoracic Society (Session A102. EVALUATING THE INTERSECTION BETWEEN AUTOIMMUNITY, IMMUNODEFICIENCY, AND INTERSTITIAL LUNG DISEASES) Abstract.
Eosinophilic granulomatosis with polyangiitis (EGPA) is a rare anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis characterized by eosinophilia and severe asthma. We conducted a Genome-wide Association Study (GWAS) of EGPA in a total of 928 cases and 11,266 controls of European ancestry. We present our findings at the annual meeting of American Thoracic Society (Session A102. EVALUATING THE INTERSECTION BETWEEN AUTOIMMUNITY, IMMUNODEFICIENCY, AND INTERSTITIAL LUNG DISEASES) Abstract.
Precision medicine has potential for transforming the healthcare delivery by more effectively matching disease screening and treatment to patient’s genetic profile. Despite many successful examples, more widespread adoption of precision medicine is hampered by the complex genetic nature of most disorders, including monogenic disorders. Here, we address the methodological gaps posed by the estimation penetrance for monogenic variants. Sung will present a liability threshold model-based penetrance estimator in Polygenic Risk Scores session.
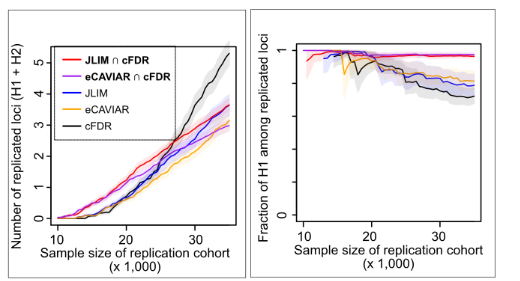 Small-scale studies often have inadequate power to discover new genetic associations. Traits that are burdensome or expensive to phenotype, rare diseases that are hard to sample and diseases that affect under-represented populations could all lead to such underpowered genetic studies. Here, we present a strategy to leverage pleiotropy between traits to discover new loci in underpowered traits. Specifically, we employ an approach that we call “consensus pleiotropy,” which combines a colocalization test with a locus-level test of pleiotropy. In simulations, we show that this approach is highly selective for identifying true pleiotropy driven by the same causative variant, thereby improves the chance to replicate the associations in underpowered validation cohorts. Using Obstructive Sleep Apnea (OSA) as an examplar, we show that we can identify and replicate a novel OSA-association in PRIM1 (DNA primase subunit 1) locus in small genetic cohorts. See Chun and Akle et al. PLOS Genet 2022 for more details of our work.
Small-scale studies often have inadequate power to discover new genetic associations. Traits that are burdensome or expensive to phenotype, rare diseases that are hard to sample and diseases that affect under-represented populations could all lead to such underpowered genetic studies. Here, we present a strategy to leverage pleiotropy between traits to discover new loci in underpowered traits. Specifically, we employ an approach that we call “consensus pleiotropy,” which combines a colocalization test with a locus-level test of pleiotropy. In simulations, we show that this approach is highly selective for identifying true pleiotropy driven by the same causative variant, thereby improves the chance to replicate the associations in underpowered validation cohorts. Using Obstructive Sleep Apnea (OSA) as an examplar, we show that we can identify and replicate a novel OSA-association in PRIM1 (DNA primase subunit 1) locus in small genetic cohorts. See Chun and Akle et al. PLOS Genet 2022 for more details of our work.
Polygenic risk scores (PRS) have potential clinical utility for risk surveillance, prevention and personalized medicine. To assess the current state of the field and stimulate the cross-polination of new ideas, we organized a community challenge for polygenic risk prediction as part of Critical Assessment of Genome Interpretation (CAGI) 6 program. Sung served as a Co-assessor to this challenge and will discuss the findings from the CAGI6 PRS challenge at the annual meeting of ASHG (Session S09, Invited Talk 053). Slides are available here.
 Eosinophilic granulomatosis with polyangiitis (EGPA) is a rare anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis characterized by eosinophilia and severe asthma. We conducted a Genome-wide Association Study (GWAS) of EGPA in a total of 928 cases and 11,266 controls of European ancestry. We present our findings at the annual meeting of American Thoracic Society (Session A102. EVALUATING THE INTERSECTION BETWEEN AUTOIMMUNITY, IMMUNODEFICIENCY, AND INTERSTITIAL LUNG DISEASES) Abstract.
Eosinophilic granulomatosis with polyangiitis (EGPA) is a rare anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis characterized by eosinophilia and severe asthma. We conducted a Genome-wide Association Study (GWAS) of EGPA in a total of 928 cases and 11,266 controls of European ancestry. We present our findings at the annual meeting of American Thoracic Society (Session A102. EVALUATING THE INTERSECTION BETWEEN AUTOIMMUNITY, IMMUNODEFICIENCY, AND INTERSTITIAL LUNG DISEASES) Abstract. Small-scale studies often have inadequate power to discover new genetic associations. Traits that are burdensome or expensive to phenotype, rare diseases that are hard to sample and diseases that affect under-represented populations could all lead to such underpowered genetic studies. Here, we present a strategy to leverage pleiotropy between traits to discover new loci in underpowered traits. Specifically, we employ an approach that we call “consensus pleiotropy,” which combines a colocalization test with a locus-level test of pleiotropy. In simulations, we show that this approach is highly selective for identifying true pleiotropy driven by the same causative variant, thereby improves the chance to replicate the associations in underpowered validation cohorts. Using Obstructive Sleep Apnea (OSA) as an examplar, we show that we can identify and replicate a novel OSA-association in PRIM1 (DNA primase subunit 1) locus in small genetic cohorts. See
Small-scale studies often have inadequate power to discover new genetic associations. Traits that are burdensome or expensive to phenotype, rare diseases that are hard to sample and diseases that affect under-represented populations could all lead to such underpowered genetic studies. Here, we present a strategy to leverage pleiotropy between traits to discover new loci in underpowered traits. Specifically, we employ an approach that we call “consensus pleiotropy,” which combines a colocalization test with a locus-level test of pleiotropy. In simulations, we show that this approach is highly selective for identifying true pleiotropy driven by the same causative variant, thereby improves the chance to replicate the associations in underpowered validation cohorts. Using Obstructive Sleep Apnea (OSA) as an examplar, we show that we can identify and replicate a novel OSA-association in PRIM1 (DNA primase subunit 1) locus in small genetic cohorts. See